DNA Methylation
Overview
Whole Genome Bisulfite Sequencing (WGBS) and Reduced Representation Bisulfite Sequencing (RRBS) are next-generation sequencing (NGS) application that profile the DNA methylation patterns genome-wide at a single-base resolution to help understand its influence on gene expression.
General Workflow
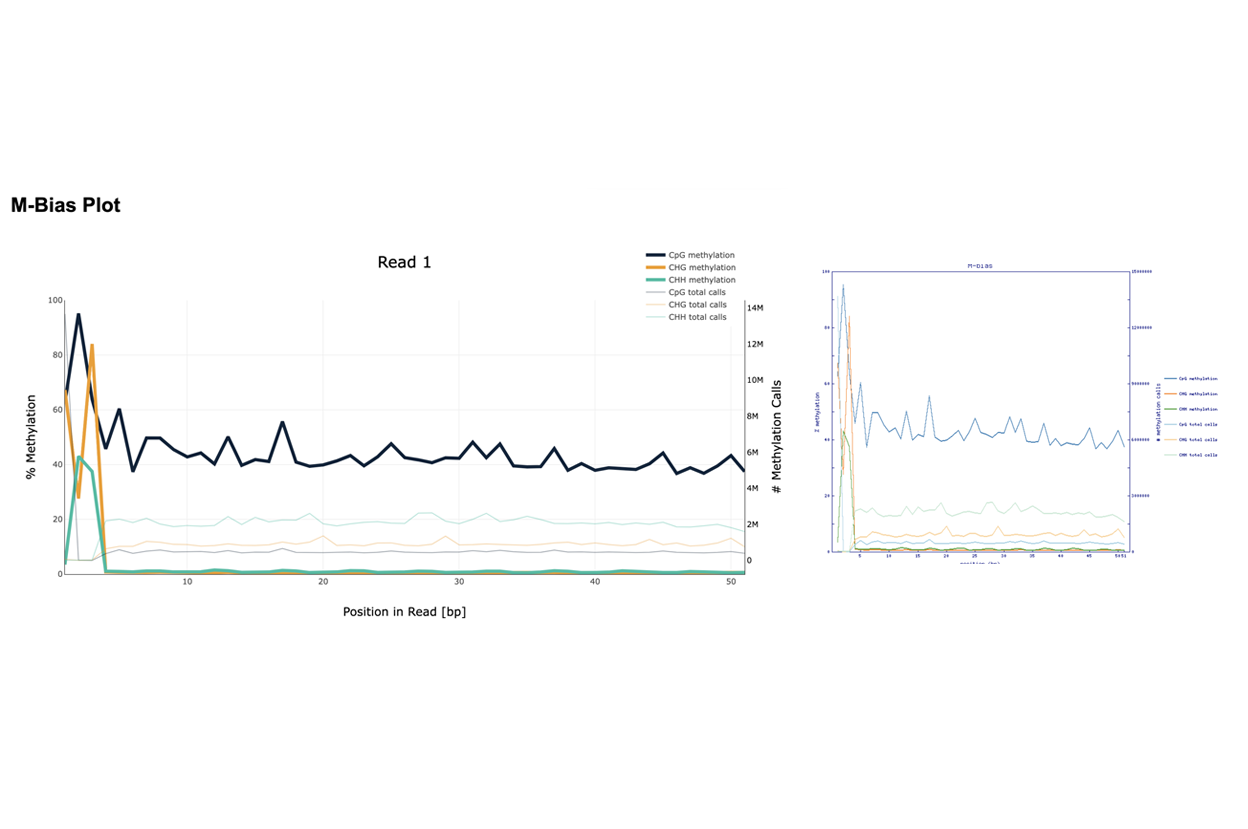
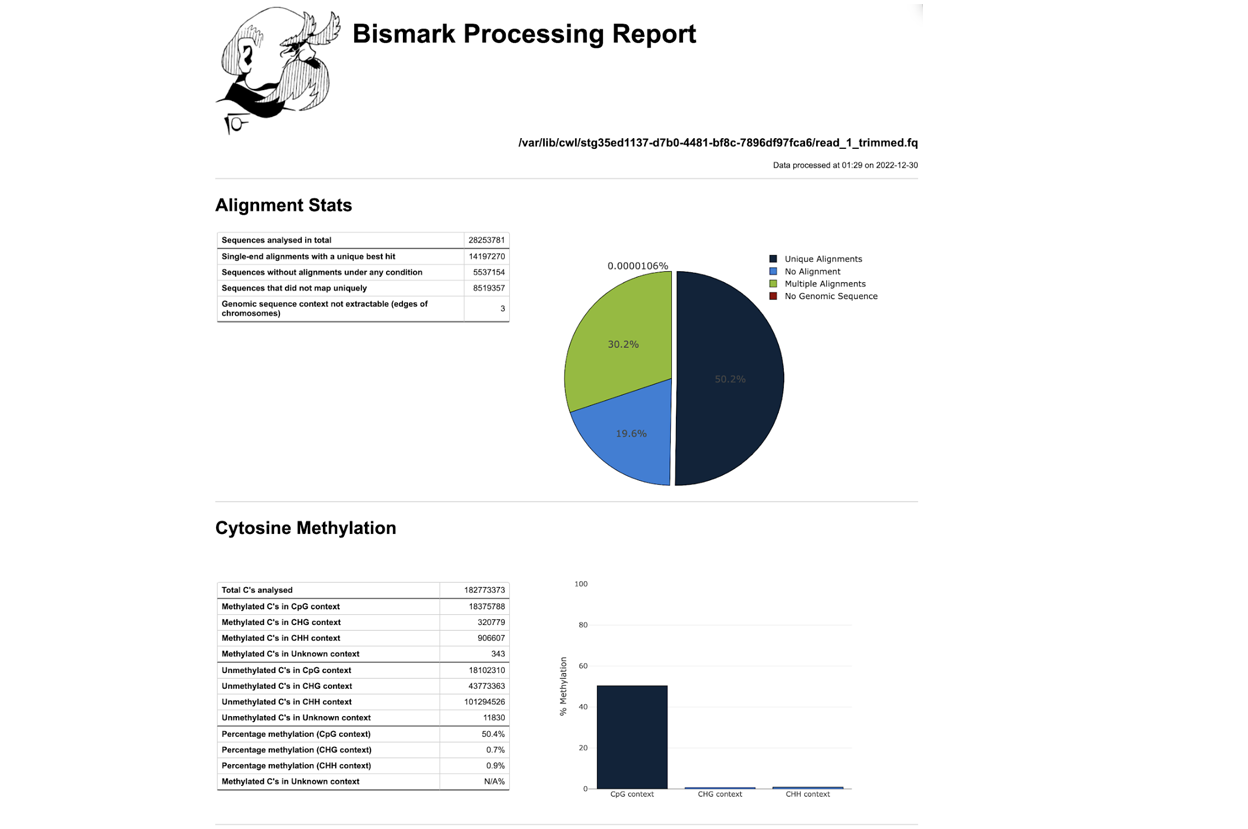
DNA methylation utilizes bisulfite sequencing to detect methylated cytosines in genomic DNA. In this method, genomic DNA is treated with bisulfite. After treatment unmethylated Cytosines (C) will be converted into Thymidines (T), whereas methylated and hydroxymethylated Cs will stay Cs. DNA is then cloned into a library, amplified and subjected to NGS.
Data Analysis
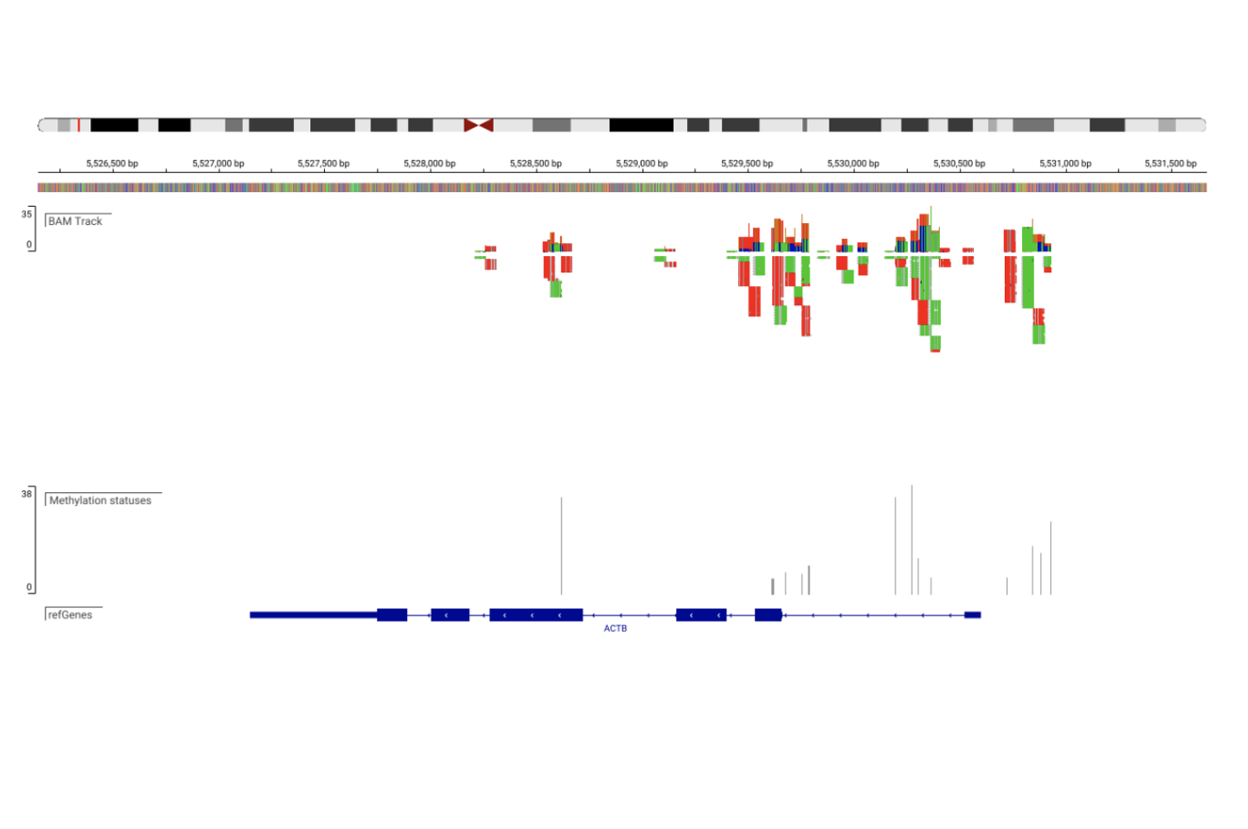
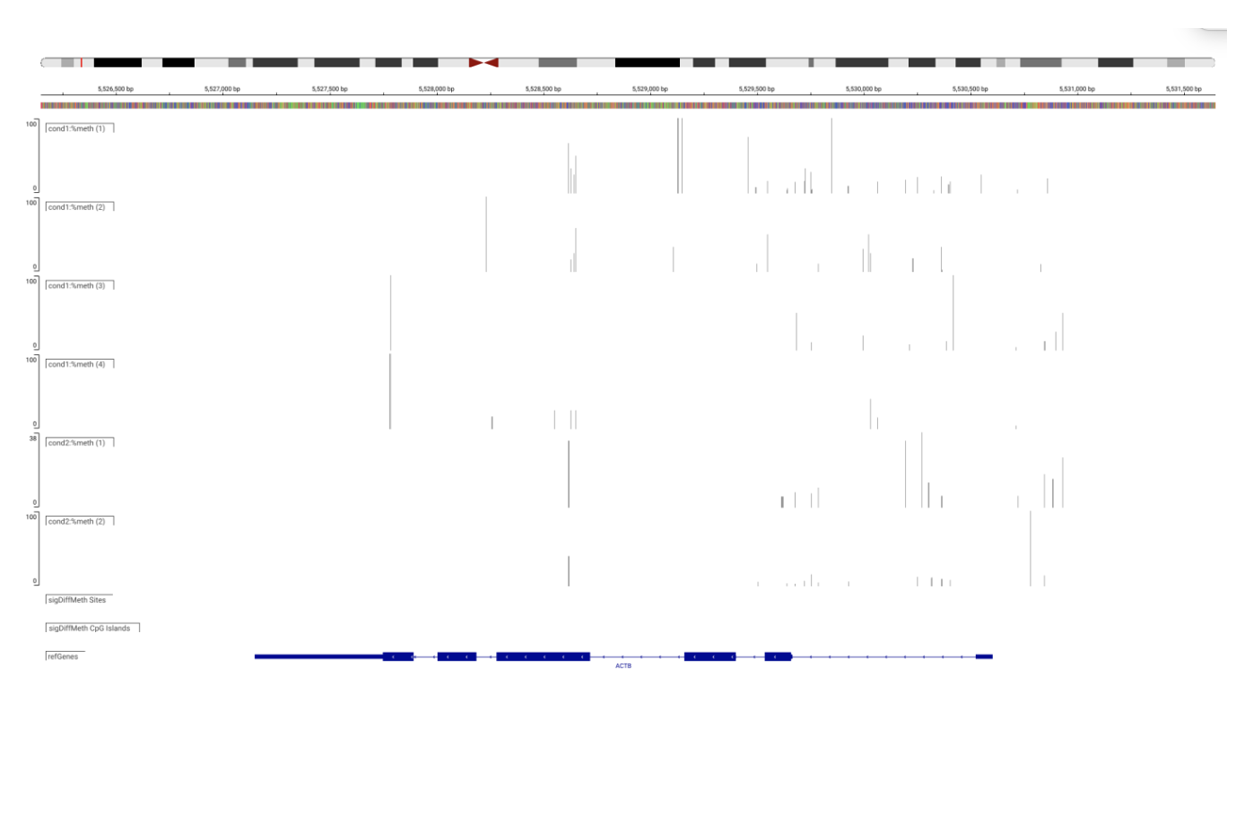
SciDAP is a no-code bioinformatics platform that enables biologists to analyze NGS-based data without a bioinformatician. It has built-in pipelines based on open-source workflows to analyze DNA methylation. SciDAP starts from the FASTQ files provided by most DNA core facilities and commercial service providers. Starting from raw data allows SciDAP to ensure that all experiments have been processed in the same way and simplifies the deposition of data to GEO upon publication. The data can be uploaded from the end-users’ computer, downloaded directly from an FTP server of the core facility by providing a URL, or from GEO by providing SRA accession number.
 scidap
scidap