Single Cell ATAC-Seq
Overview
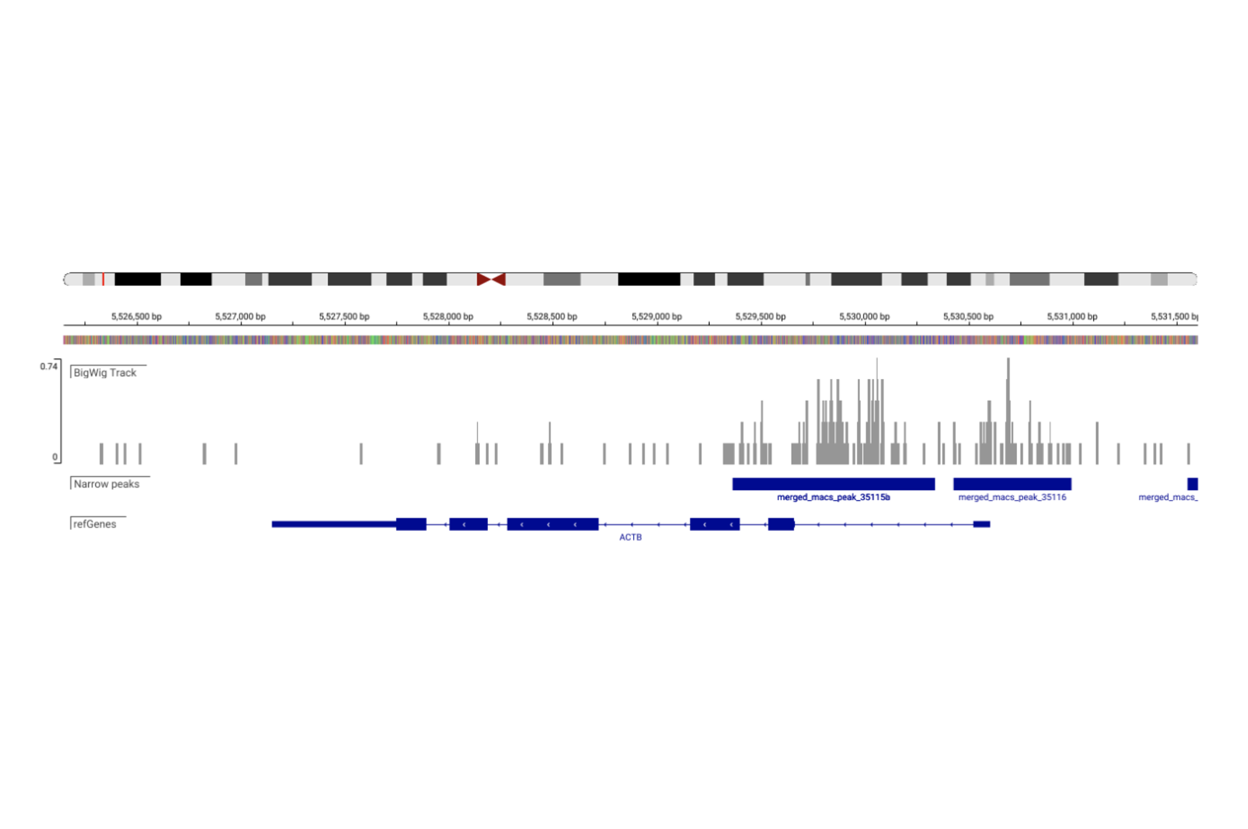
Single cell ATAC-Seq is a next-generation sequencing (NGS) application that identifies open chromatin states of individual subpopulations of cells within a sample. Open chromatin states are generally associated with sites bound by transcription factors or undergoing active transcription.
General Workflow
A typical single cell ATAC-Seq experimental workflow involves the isolation of single intact nuclei. After the nuclei are isolated, a modified Tn5 transposase enzyme inserts sequencing adapters directly onto double stranded DNA fragments. The DNA is purified, and PCR extension and amplification is performed to create an NGS library.
Data Analysis
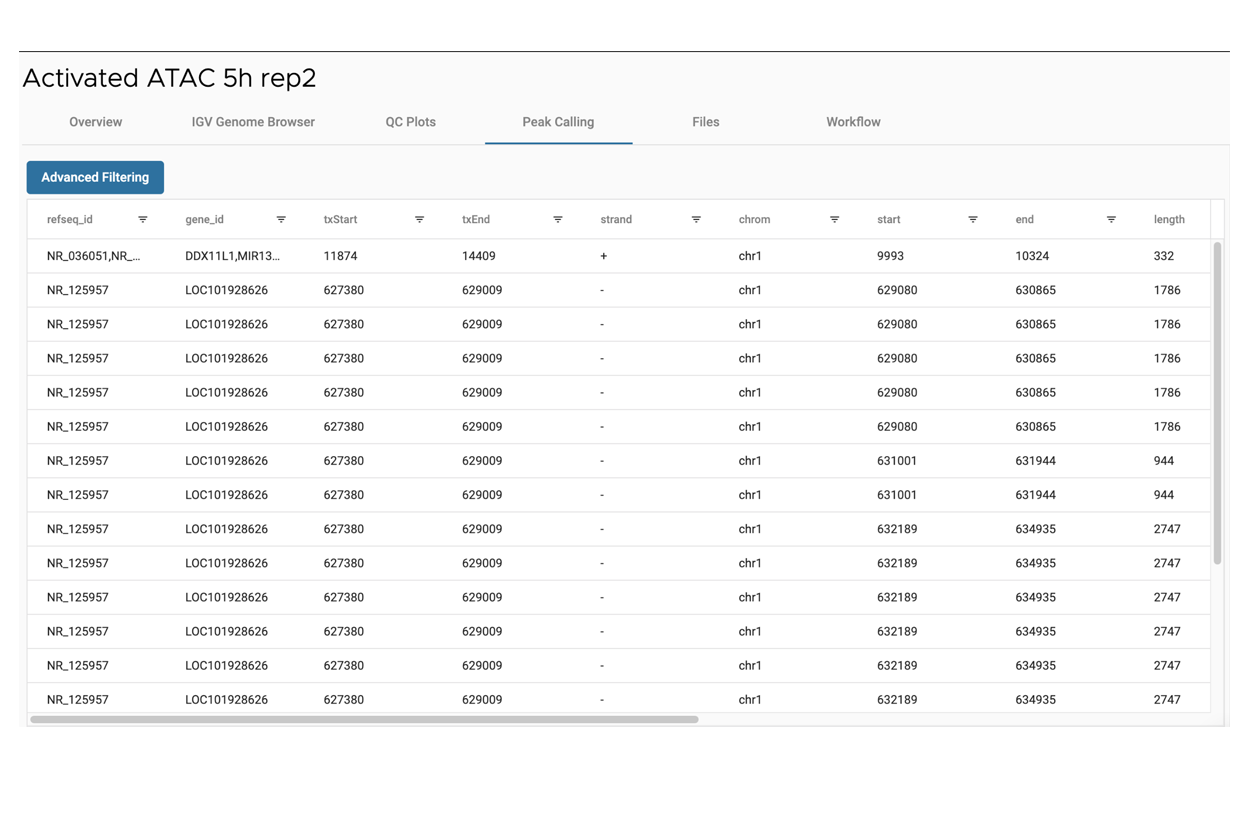
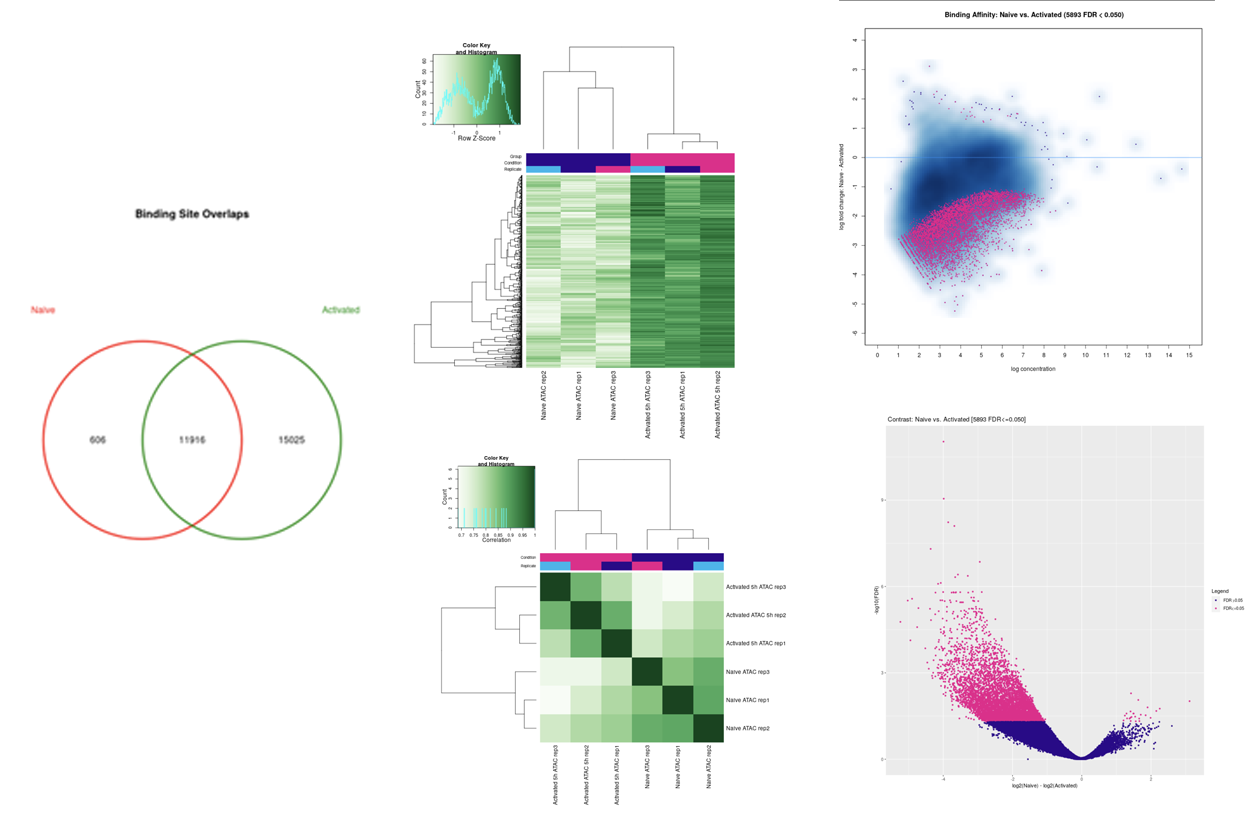
SciDAP is a no-code bioinformatics platform that enables biologists to analyze NGS-based data without a bioinformatician. It has built-in pipelines based on open-source workflows to analyze data from single cell ATAC-Seq libraries. SciDAP starts from the FASTQ files provided by most DNA core facilities and commercial service providers. Starting from raw data allows SciDAP to ensure that all experiments have been processed in the same way and simplifies the deposition of data to GEO upon publication. The data can be uploaded from the end-users’ computer, downloaded directly from an FTP server of the core facility by providing a URL, or from GEO by providing SRA accession number.
 scidap
scidap